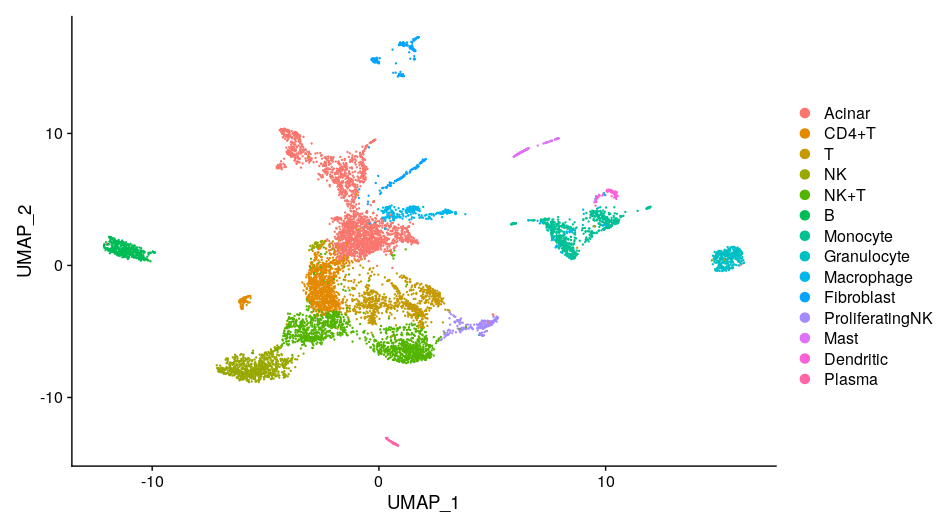
scRNA-seq
Single-cell RNA sequencing (scRNA-seq) technology is a revolutionary technique that allows scientists to understand the transcriptome at the individual cell level. This advanced method has emerged as an indispensable tool in genomics, enabling researchers to unravel the complexities of cellular diversity and function.
One of the central strengths of scRNA-seq lies in its ability to unveil the inherent heterogeneity of cellular populations. Traditional bulk RNA sequencing measures the average expression of genes across thousands to millions of cells. While it provides valuable insights, it often overlooks the contribution of individual cells, masking unique or rare cell types and states. scRNA-seq, in contrast, allows us to peek into this unseen diversity, illuminating a comprehensive landscape of cellular identities and states.
The application of scRNA-seq is wide-ranging, stretching from basic biology to medicine. For instance, in developmental biology, it can trace the trajectory of cell differentiation and the genesis of various cell types from a single progenitor. In neuroscience, it is used to delineate the vast heterogeneity of neurons and glial cells. Meanwhile, in oncology, it can uncover the tumor microenvironment, revealing not only the tumor cells’ subclonal structure but also the spectrum of immune and stromal cells involved in tumor progression and response to therapies.
Our project aims to Analysis characteristics and inhibition mechanism of tumor microenvironment (TME) through the study of interaction between pancreatic cancer and cancer associated fibroblast (CAF).